Bioinformatics Analysis and Design Based on R-studio and Databases
Feb 25, 2020· ·
0 min read
·
0 min read
Jie Zhang
Mengting Li
Abstract
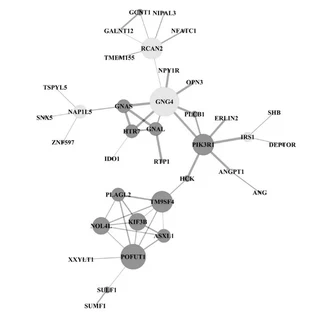
The gene chip data set with access number GSE41439 in NCBI gene expression profile database is selected as the analysis object. Firstly,the differential expression genes are screened by R-studio and the clustering heat map is drawn,then the differential genes are uploaded to DAVID database for GO function and KEGG pathway enrichment analysis,and then the protein interaction network is constructed by using STRING database,and can be seen by using Cytoscape software to observe the relationship between protein and protein directly. Four key genes,PIK3R1,GNAS,GNAL and GNG4,were screened out by protein interaction network,which can be further discussed. This method is suitable for the research of many kinds of gene chips,and has good generalization. It can be applied to the disease-related gene chips,which can provide some help for medical diagnosis and precise treatment.
Publication
In Modern Information Technology